CASE STUDY
Public-private partnership maps COVID cases, variants at international airports

Program snapshot
- Public-private partnership with CDC and XpresCheck
- Three complementary approaches of sample collection: voluntary nasal swabbing, aircraft wastewater, and airport wastewater sampling via triturators
- Sequencing of positive samples to track variants and sub-variants arriving in the U.S.
Notable highlights
- Monitoring the spread of COVID-19 and over 30 other viruses, bacteria, and antimicrobial resistance targets
- Sequenced the first confirmed instance of BA.2 and BA.3 sub-variants in the U.S.
- Filling gaps in global biosurveillance and intelligence collection
- TGS is a flexible, multimodal platform that plays an important role in U.S. national biosecurity
370,000+
Participants as of October 2023
6,000
Participants per week
7
International airports in the U.S. including JFK, EWR, SFO, LAX, IAD, SEA, and BOS

Challenge
Getting ahead of the gathering storm
Biology does not respect borders. When the CDC wanted a “weather map” of COVID cases and variants arriving in the U.S., it partnered with XpresCheck and Concentric to create the first in-airport pathogen monitoring program.
The TGS program has proven to be an agile and beneficial asset to public health officials in the United States—quickly adapting to an evolving pandemic in real time since it launched in 2021.

Solution
An early warning system for incoming biological threats
Since its inception, the program has sequenced more than 14,000 samples and made the genomic data available on several public health platforms to enable further analysis. TGS is able to monitor and change focus as needed to identify priority pathogens.
The TGS program can augment global surveillance systems, especially as testing and sequencing information become limited as COVID-19 monitoring wanes.
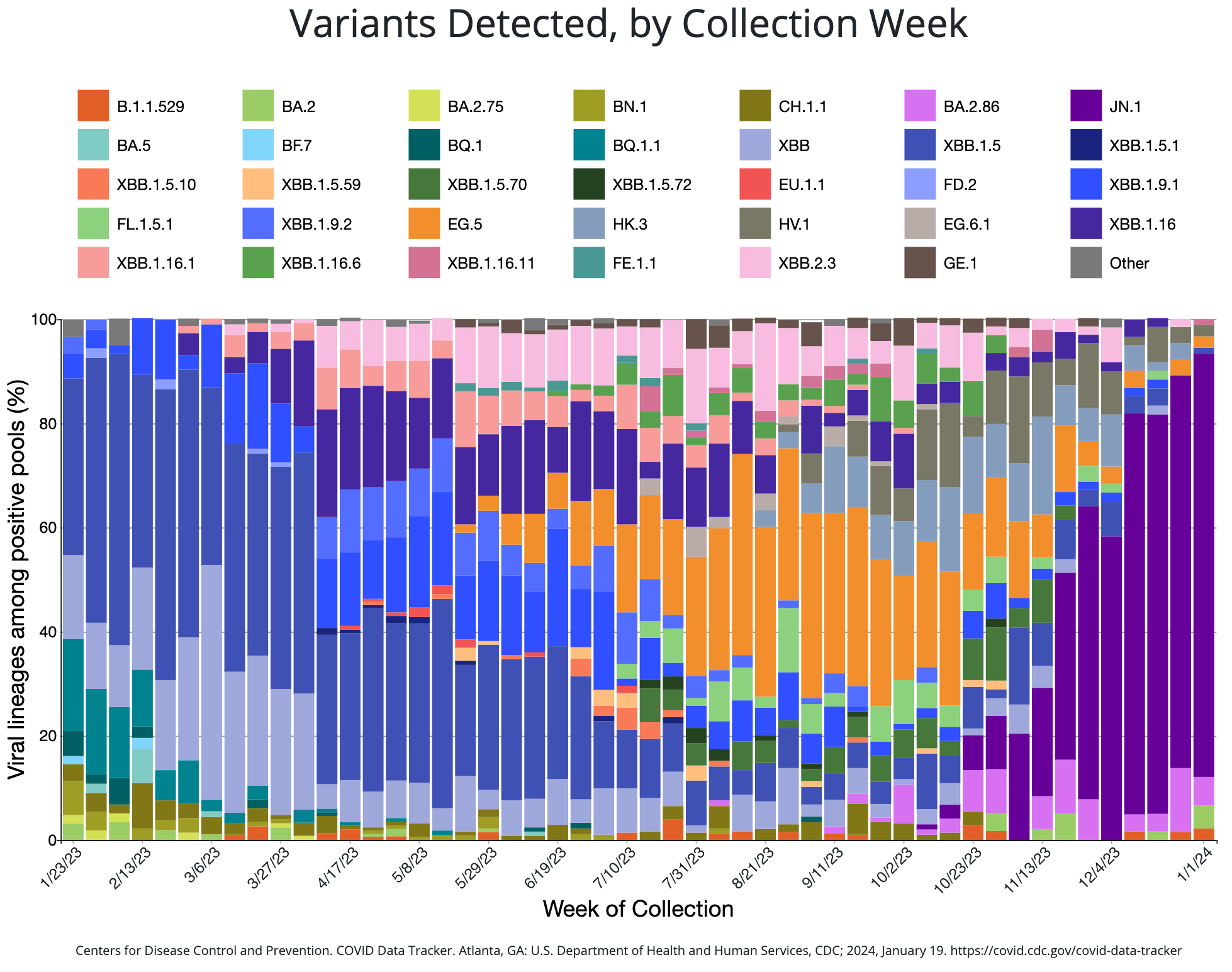
Impact
Biosecurity is national security
TGS was among the first to detect many new SARS-CoV-2 variants entering the United States up to six weeks before they were reported elsewhere, including Omicron BA2, BA3, XBB, and BA2.86 – demonstrating the program’s valuable ability to detect new variants early.
As the early warning system grows, we’ll be better prepared to respond to biological threats.